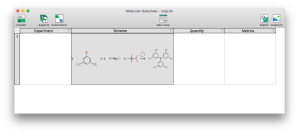
 Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term).
Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term).
Things have been moving slowly on this front on account of the fact that the lion’s share of my time has been put into advancing several time-sensitive projects for CDD (which will all be explained soon). In between that and playing a disturbing amount of Fallout 4 (what’s not to like about an immersive post-apocalyptic open world?), reaction editing on Mac OS X continues to march forward.
The fundamental datastructure is based on three layers of aspects that are in turn layered on the molecular datasheet format first used by SketchEl, then by various other products under the banner of Molecular Materials Informatics. The long story short version is that the pieces that make up a reaction description are formatted in a very regimented way which is well defined from the point of view of machine readability, which makes it suitable for electronic lab notebooks and as the grist for value added algorithms and machine learning source material that needs to analyse and understand the composition of a reaction. The Experiment layer marshals the constituent fundamental datatypes into a multistep reaction scheme with stoichiometry and quantities that can be unambiguously balanced, and various kinds of metadata. At the present time, the only complete software available for editing this data is the GLN app, which only runs on iOS mobile devices. But soon the desktop will be joining in.
Before the editor could be constructed, the datastructure had to be ported to Swift (from separate implementations in Java and Objective-C), which would have been relatively straightforward, except that the XMDS framework for editing datasheets with type-specific aspect plugins uses an awkward method for modifying the data. While the underlying datasheet is just a table of rows and columns, all modifications have to be expressed as a list of primitive operations (set cell, insert column, delete row, etc.). This is for efficiency purposes, and to enable a long sequence of undo/redo operations to be stored, without storing multiple copies of datasheets – which may ultimately grow to include collections with millions of rows, sometime in the future when not-all-in-memory support is added. These operations have to be ordered and commutative, i.e. they can be reversed to preserve the undo/redo sequence, and batched into blocks that represent a distinct editing operation. In principle there’s nothing too complex about the idea, but in practice it makes the software engineering quite fiddly, but it’s well worth it.
The next item was to port the arrange/draw sequence, which is moderately complex, but is not particularly different to the previous implementations. Deciding how to best layout a multistep reaction with arbitrary numbers of reactants, reagents and products, with the option to switch to vertical stacking, with optional constraints to a predefined boundary, requires a fair bit of thought be put into the design.
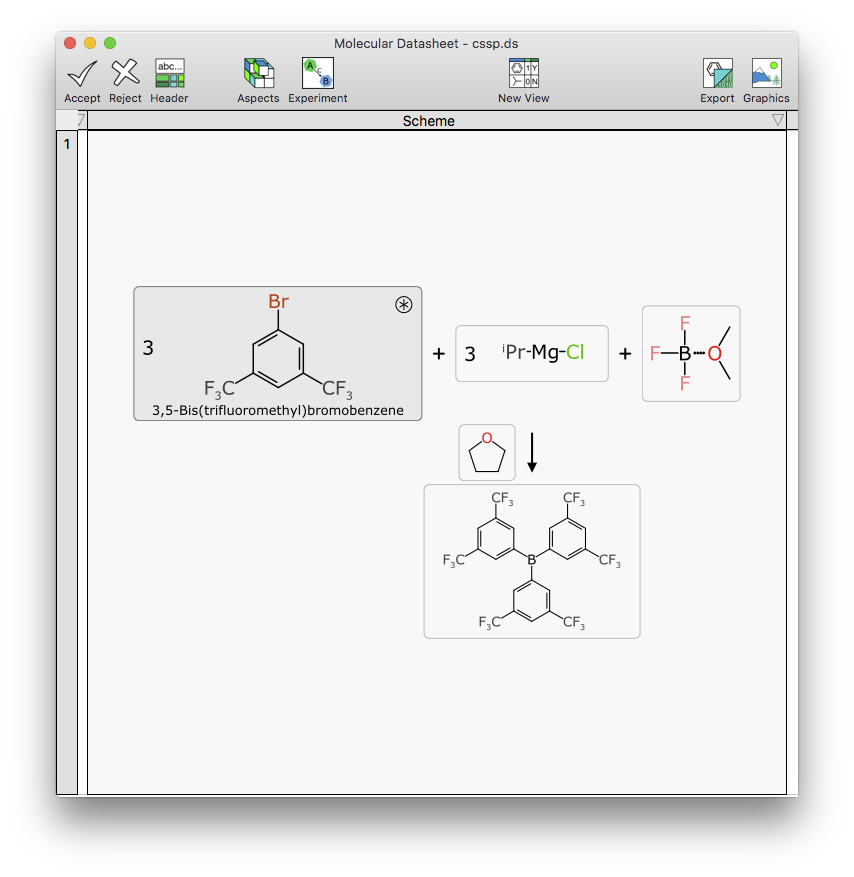
Making the components editable has two major parts. One is coopting the molecule sketcher for drawing structures, which is complicated by the very close integration between the dual spreadsheet/structure editing mode that is integral to making XMDS an ergonomic productivity tool. Making it into a three layer spreadsheet/experiment/structure editing sequence, all in the same window, sharing the same menu bar, reusing some of the same action keys, etc., takes some effort.
Editing the scalar properties of a component (name, quantity, stoichiometry, etc.) is another task that seems simple, i.e. making up a dialog box with a bunch of data entry fields. But rather than just going with a crude content capture form, it dynamically updates all of the inter-related quantities, and makes sure all the units and dependencies are properly sync’d up:
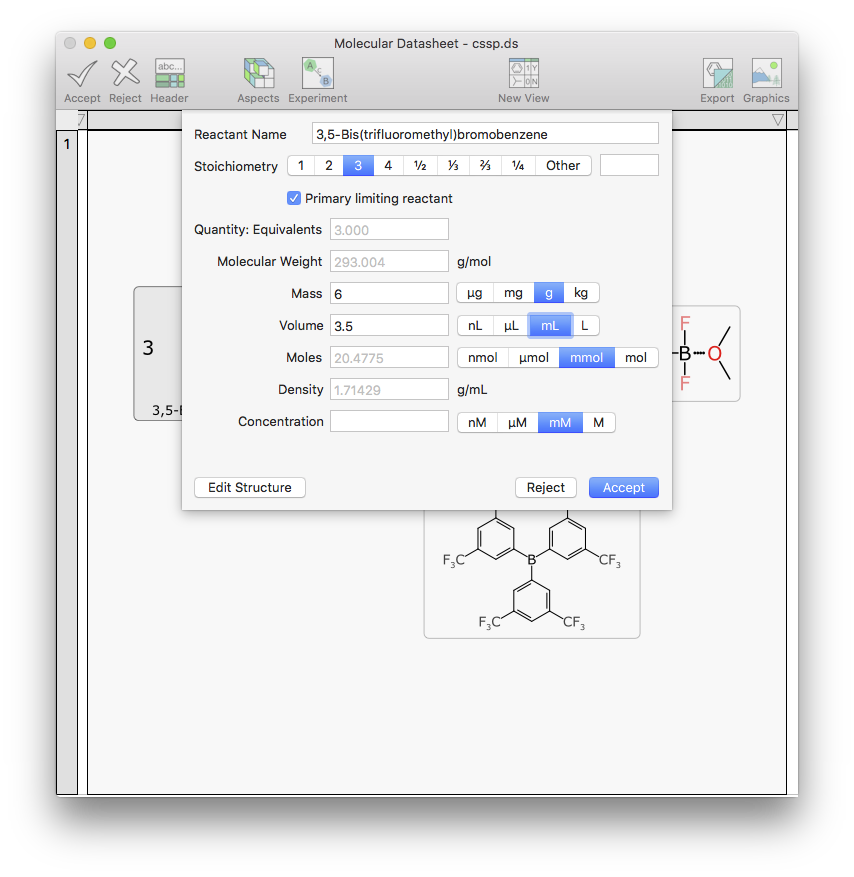
In this case, the quantities that are automatically calculated are rendered in grey, using the default value property for a text box, i.e. type over it with real data, and it is considered to be an actual experimental observation rather than a dependent calculation.
When the Experiment-containing datasheet is being displayed not in editing mode, the aspect manufactures 4 different meta-columns:
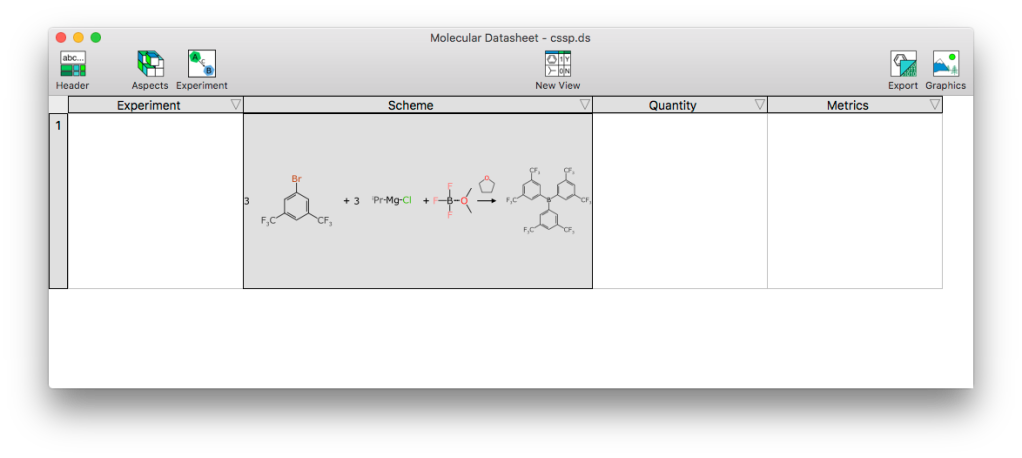
The most fundamental is Scheme, which is what this post is describing. The Experiment block contains metadata that applies to the whole experiment, which is currently just a handful of fields (title, create/modify dates and DOI). The Quantity block will display a summary the component quantities, and the editor will be have some overlap with the quantity editing for schemes, but focus on the interrelations between them. The Metrics will display certain derived properties of the reaction, specifically green chemistry properties, analogous to the GLN block.
At some point in the not too distant future, the beta test version will be updated, so you can try out the experiment content creation tools. After some more dogfooding, of course.