Next week I will be presenting at the American Chemical Society spring meeting in San Diego (2025).
What: Mixing small molecules and macromolecules in the world of sketchers
When: Tuesday 25 March, 2:55pm
Where: San Diego Convention Center Hall B-1, Room 4
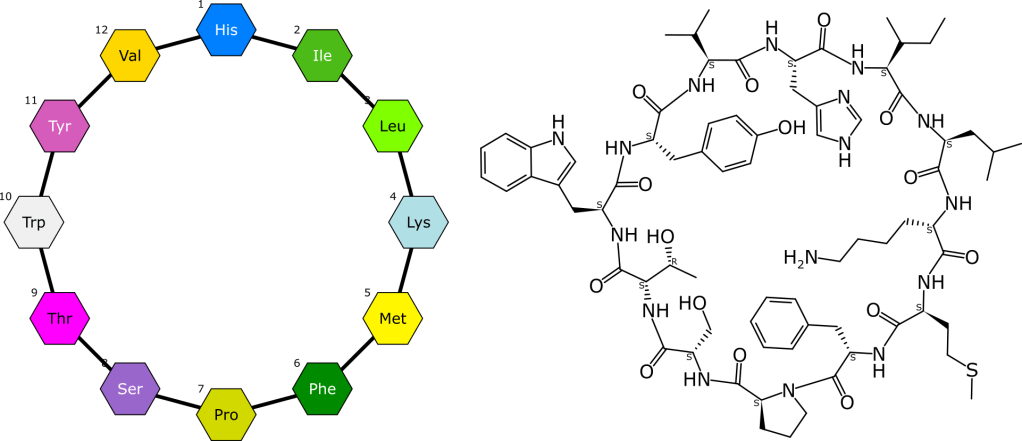
The talk will be about a major new feature that we’ve been working on at Collaborative Drug Discovery, to do with representing macromolecules as monomers embedded within a conventional connection table. It turns out that the V3000 Molfile format has an almost unknown feature for appending a list of structure templates at the end of the definition, and these can be referenced from within the main atom list. This directly leads to the ability to draw out a “cartoon” representation of a biopolymer, and draw it however you want it (i.e. it keeps the coordinates and provides various other kinds of fine grained control). Synthetic monomers can be created as necessary, or chemical structure fragments can be drawn and grafted onto monomer attachment points to create exotic hybrids. It’s also quite scalable because each template is stored just once, and because they are all stashed within the main structure, it doesn’t need an external dictionary. The full all-atom molecular structure is implied by the representation and can be expanded out losslessly.
This project began with major improvements to the open source Ketcher tool, was followed up with integration and additional functionality in CDD Vault, and also code contributions to RDKit, which can be used for chemical properties and structure searching.