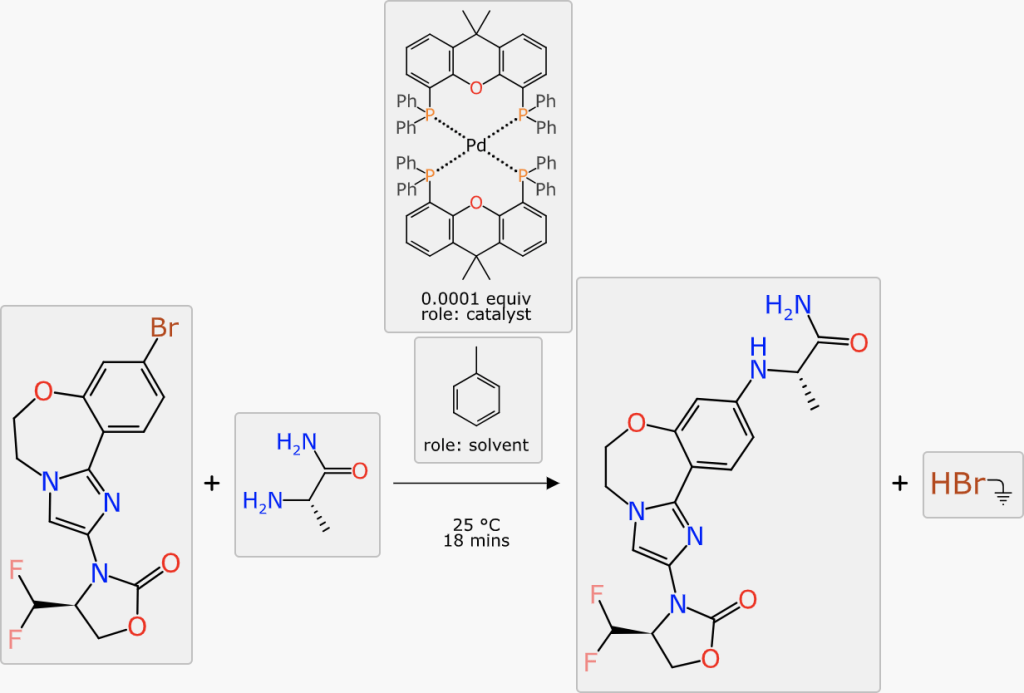
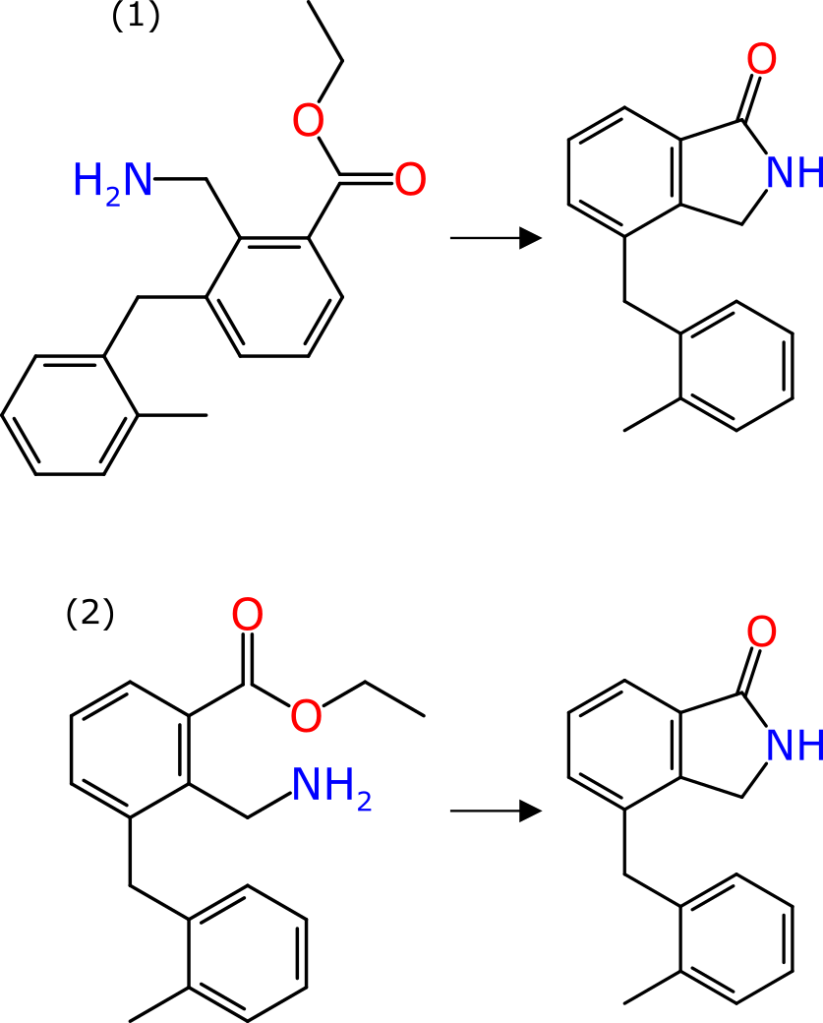
Large Language Models (LLMs or, in the parlance of our times, “AI”) have some potential value for reversing the translation of chemical reaction experiments into scientific English, into something more digitally friendly. Or put another way, there is an enormous amount of chemical reaction data that exists only as text, and if there was a less labour-intensive way to extract it, we would be much better off.
Continue readingReaction Prediction Models: Chapter 13 – Using with ELNs
Filling out content for an electronic lab notebook (ELN) is one of the main high value workflows for reaction drawing. This includes drawing the outline of a reaction that is to be performed in the lab and writing up experiments that have been completed. Either way having the reaction scheme as complete as possible, viewable by chemists and meaningful to digital archives, is important.
Continue readingReaction Prediction Models: Chapter 12 – Synthesis Planning Markup
There are a number of capable synthesis planning tools available to chemists. A retrosynthesis algorithm usually returns the barebones reactant/product pair for each step, which leaves plenty of work to do before the experiment is ready for the lab. Reaction prediction tools can help get these suggestions closer to a proposed experiment.
Continue readingReaction Prediction Models: Chapter 11 – Exporting Graphics
The reaction editor described in these articles can read & write a number of different formats, which means it does not exist in isolation. Reactions can be exported using informatics formats or as publication-ready presentation graphics.
Continue readingReaction Prediction Models: Chapter 10 – Training Data
The technology described in these articles is based on models that were built using a custom training set, or in the case of procedural algorithms, validated using that same content. The training set is unusual and proprietary, as it has been curated to a very high degree of completion and accuracy relative to any other available reaction data.
Continue readingReaction Prediction Models: Chapter 9 – Component-Based Reaction Editing
There are two main approaches to drawing reactions: the canvas model, where everything is drawn onto a flat page, and the component model, where each reaction participant is treated as a discrete object. Both of these approaches have important advantages, and significant problems which have to be addressed.
Continue readingReaction Prediction Models: Chapter 8 – Aligned Depiction
Sometimes the world of informatics overlaps strongly with the universe of human comprehension, and reaction component alignment is one of these cases: when reactants and products are drawn with a common orientation, it can be made very easy and immediately apparent to anyone with basic chemistry experience what is going on in the reaction. An arbitrary molecule layout on the other hand can impose a fairly high cognitive burden.
Continue readingReaction Prediction Models: Chapter 7 – Backward/Forward Synthesis and Reagents
Using models to propose recommendations for chemical reactions is appropriate for many of the steps needed to fill out the scheme, but sometimes it’s more effective to pick an existing reaction and use it as a template to build out the missing pieces. This approach can be applied to forward and backward syntheses (starting from a reactant or product respectively) and also to finding and proposing stoichiometric reagents.
Continue readingReaction Prediction Models: Chapter 6 – Yield Contours
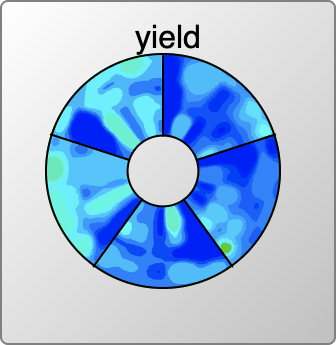
Once the reaction scheme components are all present and marked up, the last step in this toolchain is to propose a coarse-grained set of conditions, namely the duration, temperature and catalyst concentration that is likely to provide the best yield.
Continue readingReaction Prediction Models: Chapter 5 – Literature
Sometimes the best way to validate a prediction is to go check the literature and look for the most similar examples, to see if it looks reasonable. Fortunately the data that these reaction prediction tools are based only mostly comes with a DOI attached, so this feature can be added automagically.
Continue reading