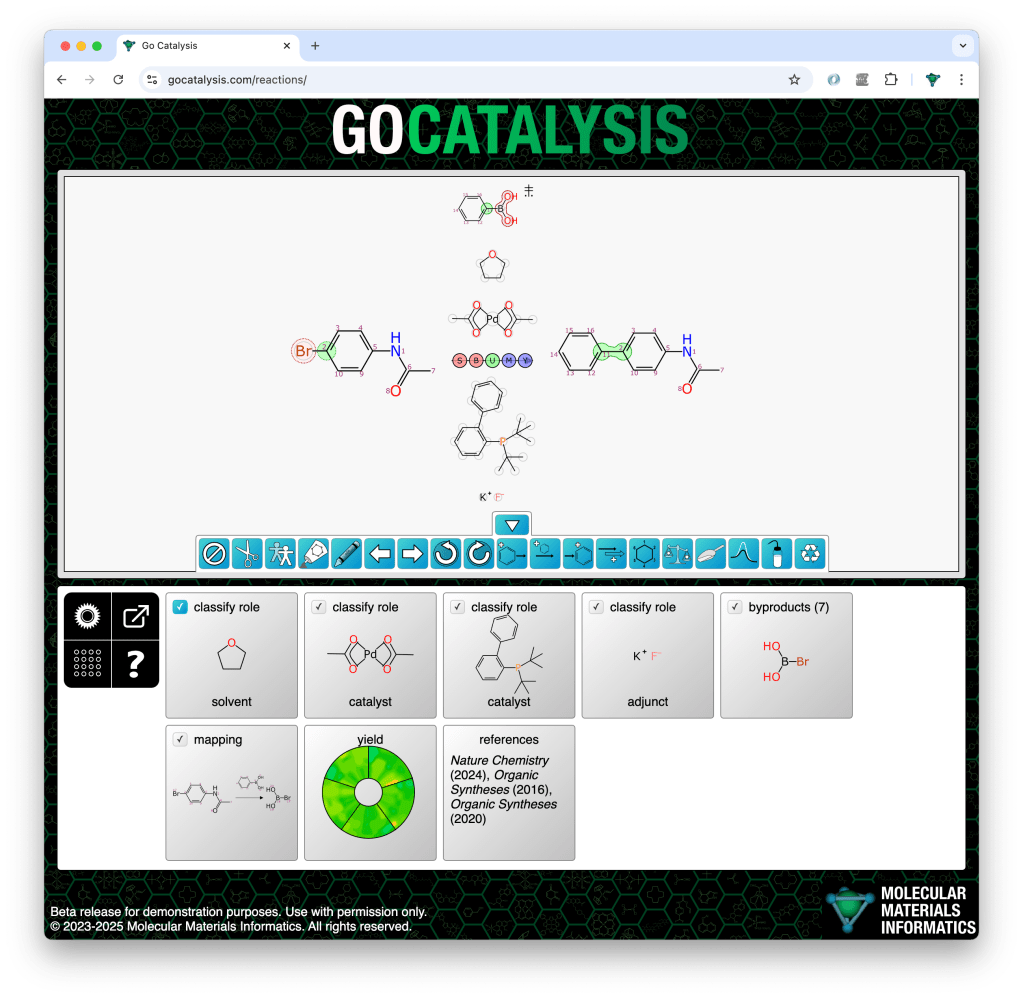
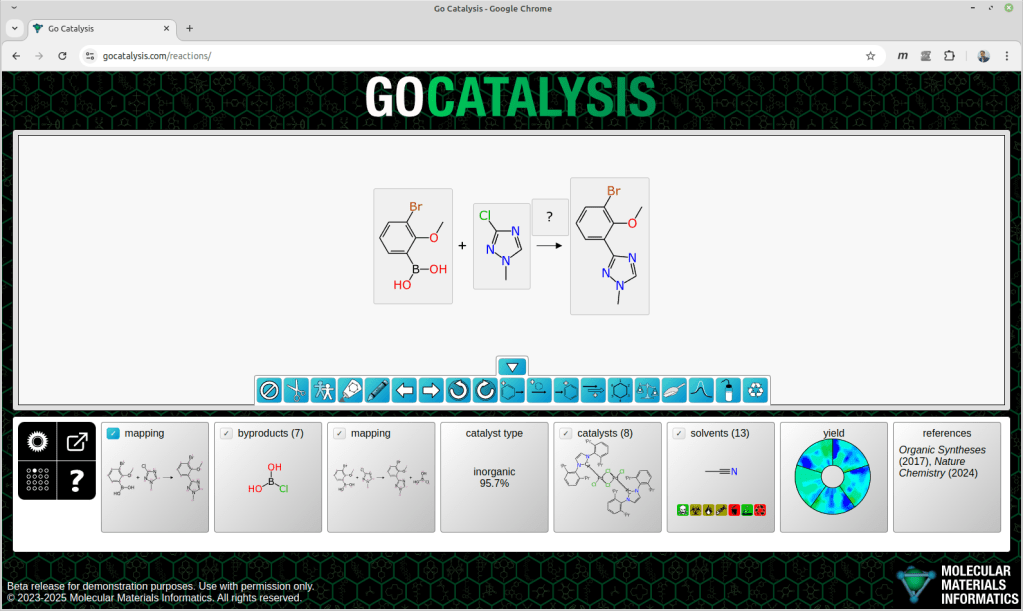
The process of completing a reaction scheme includes four preliminary co-dependent steps: proposing formal byproducts, obtaining pairwise atom-to-atom mappings, balancing the stoichiometry, and assigning roles to each of the reaction components.
Continue readingAuthor: Dr. Alex M. Clark
Reaction Prediction Models: Chapter 3 – Confidence
Prediction models for proposing and ranking catalysts and solvents are all very well, but some predictions are more reliable than others. Coming up with some kind of metric for evaluating the difference is a major improvement to utility.
Continue readingReaction Prediction Models: Chapter 2 – Solvents
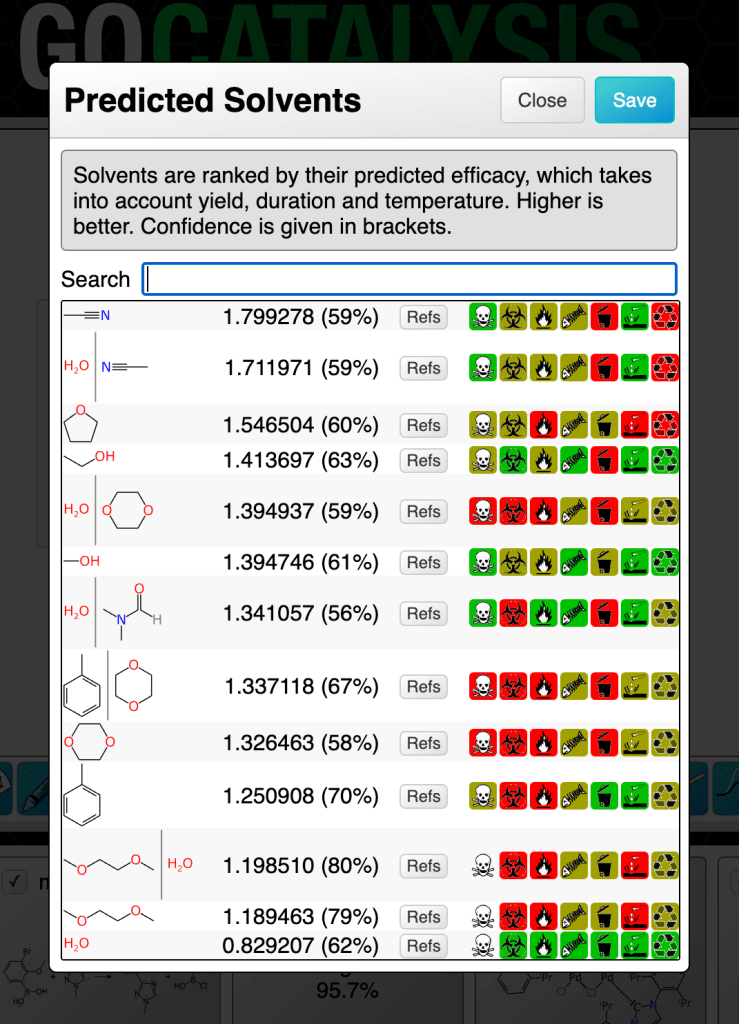
In this chapter we will explore models that can propose and rank solvents for a partially specified reaction. The methodology uses graph-based deep learning models trained on a moderate sized corpus of very well curated reactions with each of the solvents represented as a chemical structure.
Continue readingReaction Prediction Models: Chapter 1 – Catalysts
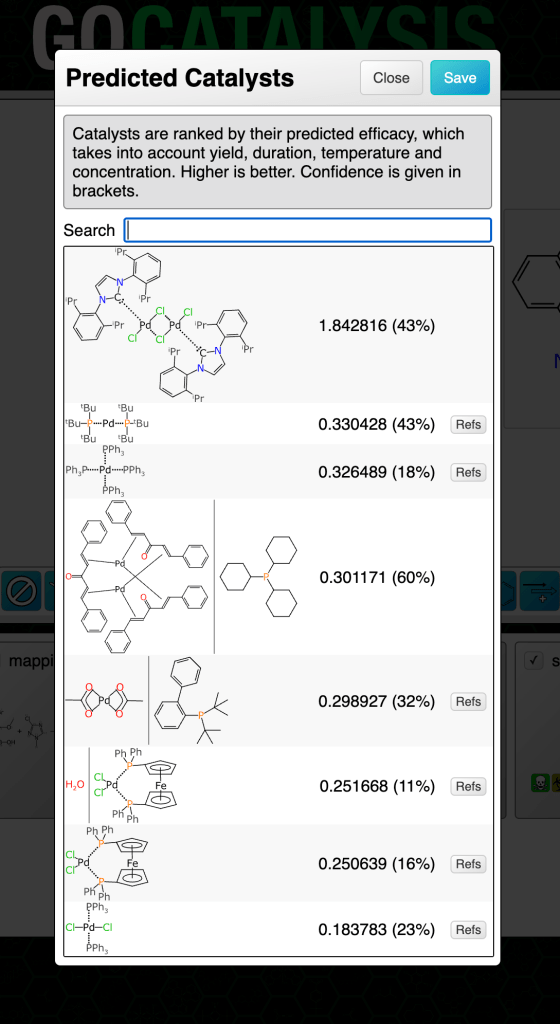
In this chapter we will explore models that can propose and rank catalysts for a given reaction transform. The methodology uses graph-based deep learning models trained on a moderate sized corpus of very well curated reactions, each of which has the catalyst molecule (or set of molecules) drawn with a chemically meaningful structure.
Continue readingReaction Prediction Models: Chapter 0
This is the first article in a series about chemical reaction prediction, in particular a work-in-progress site that combines a number of original tools for designing reactions. The general idea is that you start with an incomplete reaction scheme, and the models and algorithms will guide you toward filling out the rest, so you end up with a useful starting point for an actual experiment.
Continue readingGetting ready for ACS San Diego
Next week I will be presenting at the American Chemical Society spring meeting in San Diego (2025).
What: Mixing small molecules and macromolecules in the world of sketchers
When: Tuesday 25 March, 2:55pm
Where: San Diego Convention Center Hall B-1, Room 4
Continue readingWebMolKit 2.0 on GitHub and NPM
A new branch of the WebMolKit open source library for cheminformatics on JavaScript platforms is now available. The most noticeable change is a major source code refactor to use the modern import framework (ES6 modules), publishing with NPM, and also some new functionality such as resolving bond line crossings using a pseudo-embedding algorithm.
Continue readingOrangometallics: an inorganic structure resource
Some years ago I bought a domain name on a whim – orangometallics.com. To those of us who have dwelled within a certain chemistry subdiscipline, this is immediately obvious as a comedically simian misspelling of organometallics. Unlike the proper term, the domain name was available at a very modest price, so I decided that my company needed to own that.
Continue readingMolecule rendering: the small things
And also happy 2024 to everyone, I haven’t been writing much lately on account of juggling work and family life. The topic of this post is two aesthetic molecule drawing improvements that could be described as highly unfavourable from the point of view of the effort:reward ratio, but sometimes these things just bother you for long enough that they bubble up to the top of the to-do list.
Continue readingBioAssay Express is now open source
11 September 2023
The BioAssay Express is being released as an open source project, under the Apache 2.0 license. The short description of this license is that it is permissive, and essentially the only restriction is acknowledgment.
BioAssay Express is a grant-funded project to bring semantic web annotations to bioassay protocols, using vocabularies such as the BioAssay Ontology (BAO) to enrich descriptions that are primarily stored as text. Because of the universality of ontology terms, this means that annotated assays take on standardized meaning and can be processed by machines as effectively as they can be understood by scientists. This is a canonical example of the application of FAIR data principles (F = Findable, A = Accessible, I = Interoperable, R = Reusable).
Continue reading