 To continue on in the series of reaction-based cheminformatics on the MolSync website, the final missing search piece has now been implemented: reaction similarity. This works in a manner that is analogous to the transform feature, insofar as if you just draw one side of the reaction, or draw both sides but provide no atom-to-atom mapping information, the search behaviour just uses the molecules as-is. If you do provide atom mapping, though, the search gets a whole lot more specific. Continue reading
To continue on in the series of reaction-based cheminformatics on the MolSync website, the final missing search piece has now been implemented: reaction similarity. This works in a manner that is analogous to the transform feature, insofar as if you just draw one side of the reaction, or draw both sides but provide no atom-to-atom mapping information, the search behaviour just uses the molecules as-is. If you do provide atom mapping, though, the search gets a whole lot more specific. Continue reading
chemical reactions
MolSync reaction searching: atom mapping
 The latest unit of progress for the molsync.com site is the addition of an interface for atom mapping. As mentioned in the previous post, basic reaction searching capabilities are operational, but more sophisticated techniques require a way to specify which atoms map to each other on each side of the reaction scheme, in order to describe a transformation. Continue reading
The latest unit of progress for the molsync.com site is the addition of an interface for atom mapping. As mentioned in the previous post, basic reaction searching capabilities are operational, but more sophisticated techniques require a way to specify which atoms map to each other on each side of the reaction scheme, in order to describe a transformation. Continue reading
MolSync reaction searching: basic pieces in place
 The last update of the MolSync site added a visual overhaul and molecule searching. Today’s update brings along the basic UI for reaction searching, and preliminary functionality for various kinds of reaction searching by structure. Continue reading
The last update of the MolSync site added a visual overhaul and molecule searching. Today’s update brings along the basic UI for reaction searching, and preliminary functionality for various kinds of reaction searching by structure. Continue reading
MolSync web structure searching
 As mentioned in the previous post, the MolSync (.com) website and the technology behind it have been moving forward rapidly. The public-facing deployment now shows a proof of concept page for performing molecule searches: molsync.com/search/molecule.php. Continue reading
As mentioned in the previous post, the MolSync (.com) website and the technology behind it have been moving forward rapidly. The public-facing deployment now shows a proof of concept page for performing molecule searches: molsync.com/search/molecule.php. Continue reading
MolSync overhaul: back to the web, now with reactions too
 Things have been a bit quiet in these parts lately, but not due to inactivity: far from it. In between working on some exciting projects with Collaborative Drug Discovery, I have been quietly making rapid progress on several important key technologies. These include the OS X Molecular DataSheet (XMDS), presiding over a growing collection of reaction data, and most recently a complete overhaul of the MolSync website, which provides cheminformatics support services of various kinds. Continue reading
Things have been a bit quiet in these parts lately, but not due to inactivity: far from it. In between working on some exciting projects with Collaborative Drug Discovery, I have been quietly making rapid progress on several important key technologies. These include the OS X Molecular DataSheet (XMDS), presiding over a growing collection of reaction data, and most recently a complete overhaul of the MolSync website, which provides cheminformatics support services of various kinds. Continue reading
XMDS experiments: quantities, metrics and SVG
 The XMDS molecular datasheet editor is steadily marching forward in terms of functionality: the reaction scheme editor has been complemented by separate display panels showing the quantities of each component in blocks, which can be opened up and edited. The green chemistry metrics (PMI, E-factor, atom economy) are also calculated and shown. In the interests of making the data useful for presentations, bitmapped or vector representations can be dragged directly into a graphics package (e.g. Pages, Word, PowerPoint, Keynote, etc.), and generation of SVG images is now supported, so reactions and molecules can be easily touched-up with a vector graphics editor like Inkscape. The XMDS app (for Mac OS X) is still in long beta, so you are welcome to try it out (just ask nicely). Continue reading
The XMDS molecular datasheet editor is steadily marching forward in terms of functionality: the reaction scheme editor has been complemented by separate display panels showing the quantities of each component in blocks, which can be opened up and edited. The green chemistry metrics (PMI, E-factor, atom economy) are also calculated and shown. In the interests of making the data useful for presentations, bitmapped or vector representations can be dragged directly into a graphics package (e.g. Pages, Word, PowerPoint, Keynote, etc.), and generation of SVG images is now supported, so reactions and molecules can be easily touched-up with a vector graphics editor like Inkscape. The XMDS app (for Mac OS X) is still in long beta, so you are welcome to try it out (just ask nicely). Continue reading
Fullscreen mode in the Green Lab Notebook app
 The Green Lab Notebook (GLN) app has a little feature coming up in the next release: a mode for full screen display of an experiment, which consists of the reaction scheme and associated quantities. Continue reading
The Green Lab Notebook (GLN) app has a little feature coming up in the next release: a mode for full screen display of an experiment, which consists of the reaction scheme and associated quantities. Continue reading
Experiment editing in XMDS: dogfooding time
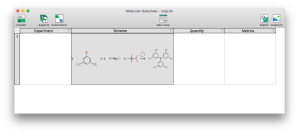 Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term). Continue reading
Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term). Continue reading