 Drawing chemical mixtures can be done online, with a conversion feature to generate Mixtures InChI (MInChI) notation. Pseudomixtures from Molfiles are now enumerated automatically when pasted in. The tools for working with machine readable mixtures, using the web platform, are open source.
Drawing chemical mixtures can be done online, with a conversion feature to generate Mixtures InChI (MInChI) notation. Pseudomixtures from Molfiles are now enumerated automatically when pasted in. The tools for working with machine readable mixtures, using the web platform, are open source.
About a year ago we published some of our work on chemical mixtures in Journal of Cheminformatics that we have been doing at Collaborative Drug Discovery. The article describes our efforts to date to define a straightforward common file format for capturing mixtures in a machine readable way, and some of the upstream & downstream use cases. The manipulation and graphical editing tools are available on GitHub: the source code is released under the Gnu Public License, while the file formats are public domain.
While you can grab the source code and tinker about with the mixture editor as much as you like, that’s not quite as convenient as clicking on a link, and so now we have one: temporarily parked at https://molmatinf.com/minchidemo. Navigating there will get you something like this:

Editing of the mixture hierarchy, components and structures can be done interactively:


The mixtures editor is coded in TypeScript, which cross compiles to JavaScript. Most of the development has been done using the Electron framework, which allows tools that are coded using the web runtime to operate as desktop apps. This provides them with many advantages over a regular web page, such as having their own window, access to local files, privileged access to resources like the clipboard, access to 3rd party web services, and the ability to execute external programs by calling operating system functions.
This last point is (or rather, was) especially important for the interaction with InChI functionality: the only reference implementation for generating InChI identifiers is a program that is written in C, and hence runs on native platforms. Most online tools that feature this technology call out to a web service, which is a bit inconvenient. Fortunately, there is a way to cross compile the InChI generator into JavaScript, and this functionality has been incorporated into the web-facing mixture editor.
What this means is that when you hit the big turquoise Create MInChI button on the demo site, some very sophisticated calculations are done all on the browser:

The colour-coded string shows the Mixtures InChI notation in its consituent parts (header, structures, hierarchy, concentration). The structure parts are made up of the main payload for each of the constituents within the mixture hierarchy.
It should be noted that the demo web app is just a bunch of files sitting on a webserver. There is no back-end, other than serving up the files (i.e. the dumbest possible HTTP server). The interactive functionality and the InChI calculation is all done on the browser itself. There are a few things that the browser incarnation can’t do, such as accessing files, but this can be partially alleviated by using the clipboard. But this means that setting up the site is a simple matter of copying the files to a web-shared directory, and the functionality operates without ever needing to send anything back to the server, which is great for both performance and security.
On the subject of clipboards, there have been some recent updates to the underlying codebase. While the Mixfile format that we made up has compelling properties for storing rich data in a community-friendly way, there are a few other ways to draw mixtures using existing tools. This includes some of the more advanced features of the Molfile CTAB format: while they may not be broadly supported, the official Biovia Draw application does facilitate several basic kinds of mixture drawings, which are be unpacked upon paste.
Advanced stereochemistry is one scenario where enumeration can be more convenient and/or intuitive to work with than adding metadata to a single reference structure. In the following example, a molecule with 3 stereocentres has been designated with each of these as part of an “OR” block, which is a way of saying that it is a mixture of the stereoisomer as-drawn, as well as its mirror image:

Pasting this structure into the mixture editor will enumerate the two valid forms:

Note that all of the other permutations were not enumerated, because the allowed options were specified precisely in the extended CTAB definition. Consider a more exotic molecular configuration, which has two of these OR-blocks:

Enumeration generates 4 outputs, since there are 2 blocks, each of which has two configurations:

The mixture importer has done the enumeration correctly, but note that it is not smart enough to spot the meso symmetry. That might be added later, but for now it is taking the user-instruction literally, for better or for worse.
Enumeration of actual structures is also possible. The brute-force way to do this is to draw each of them within the Molfile canvas, and mark them as separate components, e.g. the xylenes problem can be solved as:

Which is imported as:
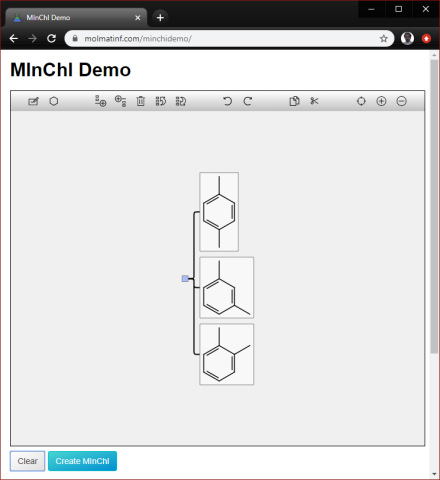
It is quite common to see databases containing a Molfile with all 3 isomers of xylene drawn side by side without any metadata (which is totally wrong) – but as long as the partitioning is provided, the importer will separate them out into components.
It is also possible to create hierarchies in this way, i.e. mixtures-within-mixtures by wrapping brackets within brackets:

This is imported as:

Note that rather than just importing 4 components into an array, it creates 2 branch nodes, each of which is populated with its two constituents.
This style of drawing mixtures is quite convenient, and may be more familiar to chemists than the hierarchy editor that comes with our mixture tools. However, there does not appear to be any official way to add extra metadata about components, such as name, concentration, etc. Some of the Biovia documentation does hint at the existence (or proposal?) of such functionality, but for now we are sticking to tools that are broadly in use. So if the mixture has information such as relative ratios or other quantity information, this needs to be added after the fact.
Another way to encode xylenes is to use the multiple attachment feature:
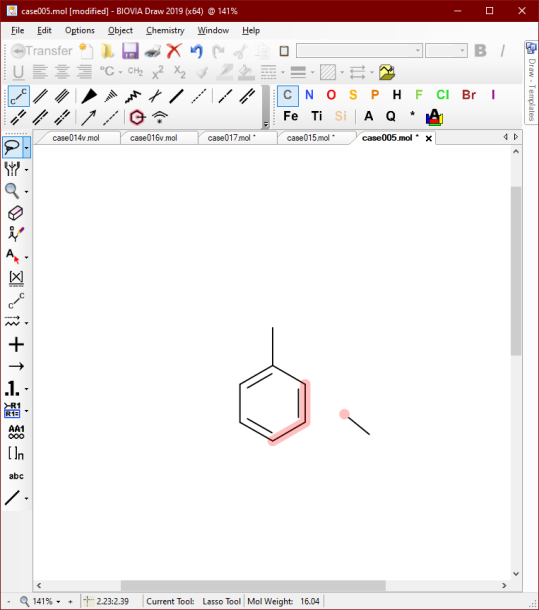
When this recipe-of-mixtures is imported, it gets expanded out to its 3 implied components:

As you can see the aesthetics are not great, since the enumeration process involves only reattaching the bond and deleting the atom placeholder. A future improvement would be to do a partial redepiction and glue on the attachment in an optimal orientation, and also check the products for degeneracy. Nonetheless, the informatics is correct and the diagram is quasi-legible, which is a fair starting point.
This is just a little preview of what’s going on with mixtures at Collaborative Drug Discovery. There is a whole lot more coming up with regard to importing unstructured text and integration with tools like the ELN, so expect to hear more over the coming months!