My latest contribution to GitHub is PDFBoxLite, which represents a few hours work. Basically, all of my software efforts that involve creating chemical graphics are leaning toward pushing PDF as the format of choice for creating inline figures. Everyone knows PDF as the Portable Document Format – things you look at before sending to the printer. But what you may not know is that PDF files with just a single page, of arbitrary size, are a very well supported as a way to get pictures into a larger document (Word, Pages, PowerPoint, Keynote, Excel, etc.). This is particularly true within the Apple ecosystem. Unlike SVG, correct rendering is essentially universal; and unlike WMF/EMF, it works on platforms other than Windows. And unlike PNG, it isn’t a trainwreck when you try to print/zoom/convert your document. Continue reading
Fullscreen mode in the Green Lab Notebook app
 The Green Lab Notebook (GLN) app has a little feature coming up in the next release: a mode for full screen display of an experiment, which consists of the reaction scheme and associated quantities. Continue reading
The Green Lab Notebook (GLN) app has a little feature coming up in the next release: a mode for full screen display of an experiment, which consists of the reaction scheme and associated quantities. Continue reading
Tox21 toxicity measurements and PolyPharma
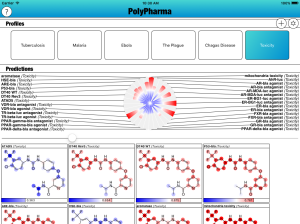 The PolyPharma app is currently getting some major new content added to it, namely a bunch of new models for toxicity. The models are derived from measurements made on the EPA’s Tox21 collection, recently published in nature, and released via PubChem. Continue reading
The PolyPharma app is currently getting some major new content added to it, namely a bunch of new models for toxicity. The models are derived from measurements made on the EPA’s Tox21 collection, recently published in nature, and released via PubChem. Continue reading
Adventures with KNIME: pipeline wrapping
 One of the items that has been on my to-do list for quite some time is to dig back into the KNIME universe, and interface it with the growing collection of cheminformatics algorithms that I have been assembling over the last few years. Given that my company’s software stack has its own workflow/pipelining infrastructure, but no user interface, it makes a certain amount of sense to look into connecting them together. Continue reading
One of the items that has been on my to-do list for quite some time is to dig back into the KNIME universe, and interface it with the growing collection of cheminformatics algorithms that I have been assembling over the last few years. Given that my company’s software stack has its own workflow/pipelining infrastructure, but no user interface, it makes a certain amount of sense to look into connecting them together. Continue reading
Panel of Bayesian screening for BIA 10-2474
The around town in drug discovery right this moment seems to be focused on BIA 10-2474, which my frequent collaborator Sean Ekins has weighed in on at the Collaborative Chemistry blog. In a spur of the moment effort to see if we could use some of our work-in-progress technologies to learn something about what’s going on, we ran it through a series of 1800 Bayesian models that we extracted from ChEMBL. For a detailed view, check out this link on molmatinf.com. The file is close to 20MB, so be patient if you’re on a slow connection. Continue reading
Composite Bayesian models: latest open source project
 My latest publication has just come out as early access in Journal of Chemical Information & Modeling, entitled “Open Source Bayesian Models: 3. Composite Models for Prediction of Binned Responses“. This is an extension of previous work on the Bayesian/fingerprint theme, and in the interim while waiting for peer review, we have some additional developments to share. Continue reading
My latest publication has just come out as early access in Journal of Chemical Information & Modeling, entitled “Open Source Bayesian Models: 3. Composite Models for Prediction of Binned Responses“. This is an extension of previous work on the Bayesian/fingerprint theme, and in the interim while waiting for peer review, we have some additional developments to share. Continue reading
Experiment editing in XMDS: dogfooding time
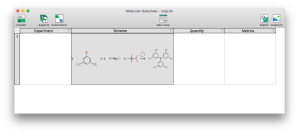 Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term). Continue reading
Progress is coming along nicely with the Experiment aspect editor for XMDS, which is essentially the desktop platform playing catchup with mobile and matching the functionality of the Green Lab Notebook (GLN) app. There is now an editor in place for the overall reaction scheme, which allows the components of a multistep reaction to be composed. This is a development milestone because it means that it’s time to switch to using it for data entry to find out what needs to be added or fixed most urgently, rather than implementing essential features that are obviously missing (cf. dogfooding = eating one’s own dogfood, in case anyone is unfamiliar with the term). Continue reading
Reactions in XMDS
 After much procrastination, chemical reactions have started to make their way into the OS X Molecular DataSheet (XMDS) beta. The screenshot shows several multistep reactions from my Ph.D. research, and were originally entered using the Green Lab Notebook (GLN) app.
After much procrastination, chemical reactions have started to make their way into the OS X Molecular DataSheet (XMDS) beta. The screenshot shows several multistep reactions from my Ph.D. research, and were originally entered using the Green Lab Notebook (GLN) app.
The Mac app and the iOS app both use the same data format, which is based on the extensible aspect protocol for molecular datasheets (see description), which seeks to capture a reaction description that is highly machine readable. As well as representing a multi-step schema, which can handle the non-organic species that are so commonly used as reactants, it also handles stoichiometry and quantities, in a way that can be used to balance the reaction and generate metrics. This has been available on mobile for awhile now, but the desktop analog is catching up.
Editing has yet to be implemented, but there’s a lot of background work that goes into just displaying reaction schemes, so it’s well underway.
Breaking radio silence
This site hasn’t had a post for couple months, and there’s a reason for that: I’ve been out of town, doing some collaborative work on-site, but now things are getting back to normal. Continue reading
MMDS with a news cycle feed (and how to turn it off)
 The latest version of the Mobile Molecular DataSheet (for iOS: see AppStore) has a news ticker feature, which shows a rotating series of facts and links that are displayed along the bottom of the screen. The items include mentions of other related apps, as well as news from Molecular Materials Informatics, and articles from this blog. The functionality is the same as what originally appeared in the free MolPrime app. Continue reading
The latest version of the Mobile Molecular DataSheet (for iOS: see AppStore) has a news ticker feature, which shows a rotating series of facts and links that are displayed along the bottom of the screen. The items include mentions of other related apps, as well as news from Molecular Materials Informatics, and articles from this blog. The functionality is the same as what originally appeared in the free MolPrime app. Continue reading