 Lately I’ve been working on a new extension to the InChI identifier which is intended to broaden its domain to include the universe of non-organic compounds and all of the insane diversity of exotic bonding types. Preliminary results of the first stage are up on GitHub.
Lately I’ve been working on a new extension to the InChI identifier which is intended to broaden its domain to include the universe of non-organic compounds and all of the insane diversity of exotic bonding types. Preliminary results of the first stage are up on GitHub.
This blog has been quiet for awhile, mainly because the day job has been quite demanding (commercialisation of the BioAssay Express product is a rather large undertaking). One of my after-hours hobby projects for the last few months has involved reconnecting with my inorganic chemistry roots, which has been enabled by the InChI Trust. The organisation shepherds the development of the InChI identifier, which is a canonical line notation which is very good at funnelling organic compounds into a unique string that can be used for disambiguation and fast database lookup. While the identifier is very effective within its domain, it grew up in an environment where cheminformatics enthusiasts were only interested in drug-like molecules, which are mostly confined to a very small set of Lewis octet-compliant bonding types. Even this small-but-critical sliver of the periodic table offers a seemingly limitless stream of edge cases to ensure that cheminformatics-of-drugs is not likely to declare victory and go looking for new problems to solve anytime soon.
Nonetheless there has lately been an increasingly loud chorus of people pointing out that having such an important suite of technologies fail hard as soon as a metal shows up is a major limitation. The primary reason that the InChI technology cannot handle metals properly is due to the deliberate decision to delete all bonds to metals during the preprocessing step. This destroys rather important information (i.e. connectivity), but in principle it would not completely eliminate the utility, because it could still be used as a hash code for prefiltering. Unfortunately the various normalisation procedures that are applied under the hood mean that this reduced condition cannot be fulfilled by a large subset of inorganic compounds (currently estimated to be ~20%).
The idea of extending the InChI identifier to work properly for inorganic compounds has been discussed for a long time, and the point at which I decided to jump in was when a formal request-for-proposal was issued, which centred around the creation of a test set to scope out the extent of the problem. This data-first approach appealed to me, in part because I already had a significant collection of handcrafted inorganic structure sketches lying around, and was also boosted by the opportunity to work with the Cambridge Structural Database (CSD) which is a foundational resource for the kinds of research that I used to do back in my graduate student days (and it even includes the structures I published).
I’ve written (and ranted) quite a lot about the tools and datastructures needed to capture inorganic compounds with the same efficacy that we expect from their simpler organic relatives, include a 2011 paper extolling the virtues of the zero-order bond. When inorganic compounds are sketched out the result is generally a hot mess, where none of the valences make sense, hydrogen counts are left unspecified, charges are poorly localised, and undefined abbreviations run rampant. With the exception of simple structures that follow the same rules as organic chemistry, it is more often than not impossible to work out the correct molecular formula, and everything goes downhill from there.
Enter the Cambridge Structural Database:
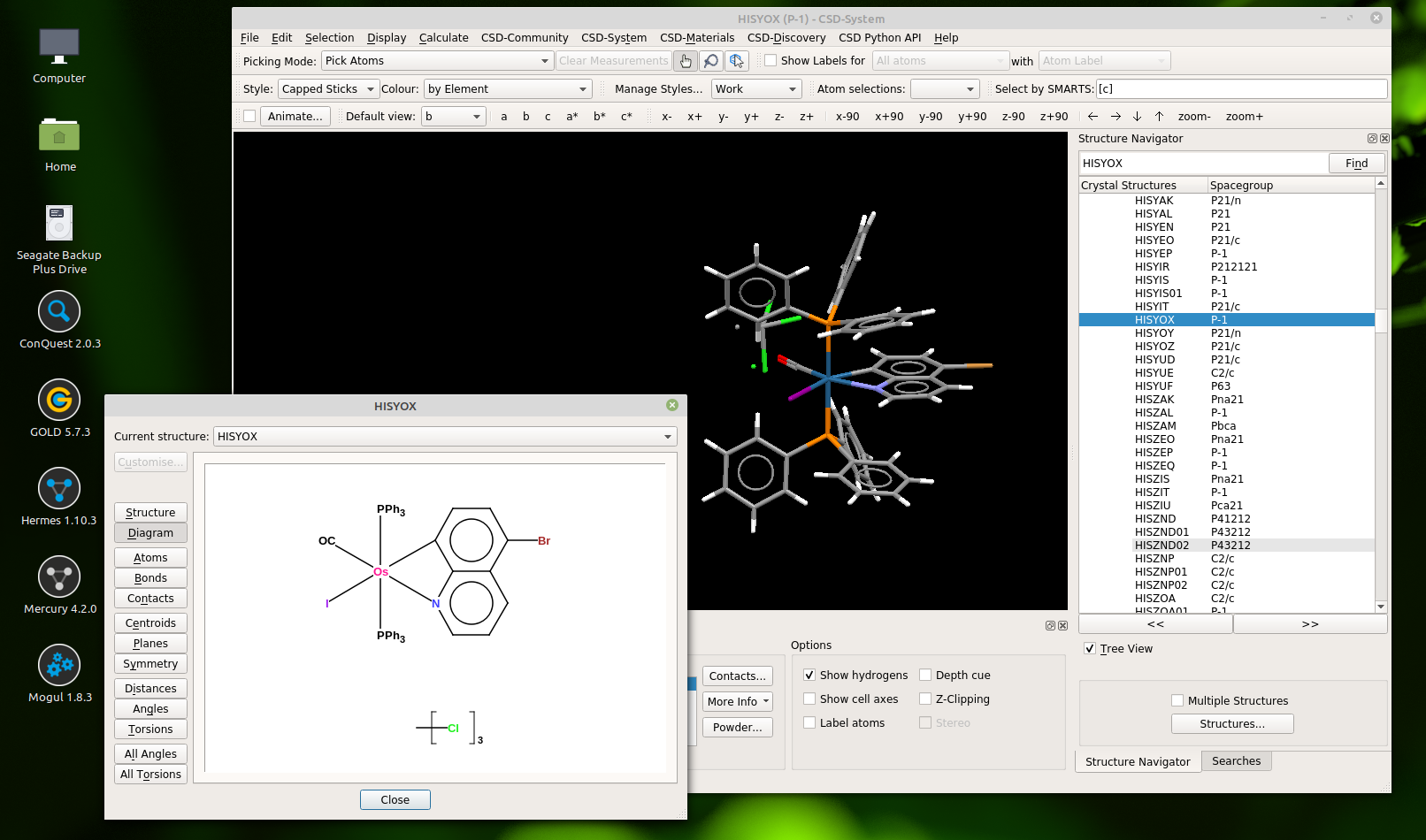
The 3D structure in the background is the usual display mode since it is representing the underlying experimental observation. The window in front of it is the exciting part, even though it looks rather drab by comparison. The 2D diagram is evidence of carefully curated information that reconciles the crystal structure solution with the interpretation of multiple domain experts.
It turns out that if you know where to look (and there’s probably no reason that you would because it’s still an undocumented secret), there is a lot more to this diagram than just the picture. The raw content contains all of the atoms, including the hydrogens (whether they were observed or not), a reasonable attempt to classify the bond types, well localised charges, and a rather good 2D layout with fully expanded abbreviations.
So this basically means that the CSD contains about 500,000 inorganic compounds with sketched coordinates and a complete graph with all atoms and bonds. This is a resource that exists in few other places, and certainly nothing on this scale. The amount of work that went into getting these details right is the result of continuous effort expended over many decades. This makes it the perfect resource to start digging into in order to find good examples for which molecules ought to work properly with a new-and-improved InChI identifier, because a huge variety of chemistry is represented, and each example is from the real world.
Back to the coordination InChI project: the details are quite thoroughly explained on the GitHub repository itself, so there’s no need to rehash in a lot of detail. The short version is that I used clustering & cherry-picking to select about 500 inorganic compounds from CSD, PubChem and my own private collection, in order to gain as much coverage as possible.
By way of illustration, these are some compounds that serve as exemplars of certain kinds of bond patterns which are commonly observed within inorganic chemistry, but not well handled by normal cheminformatics:
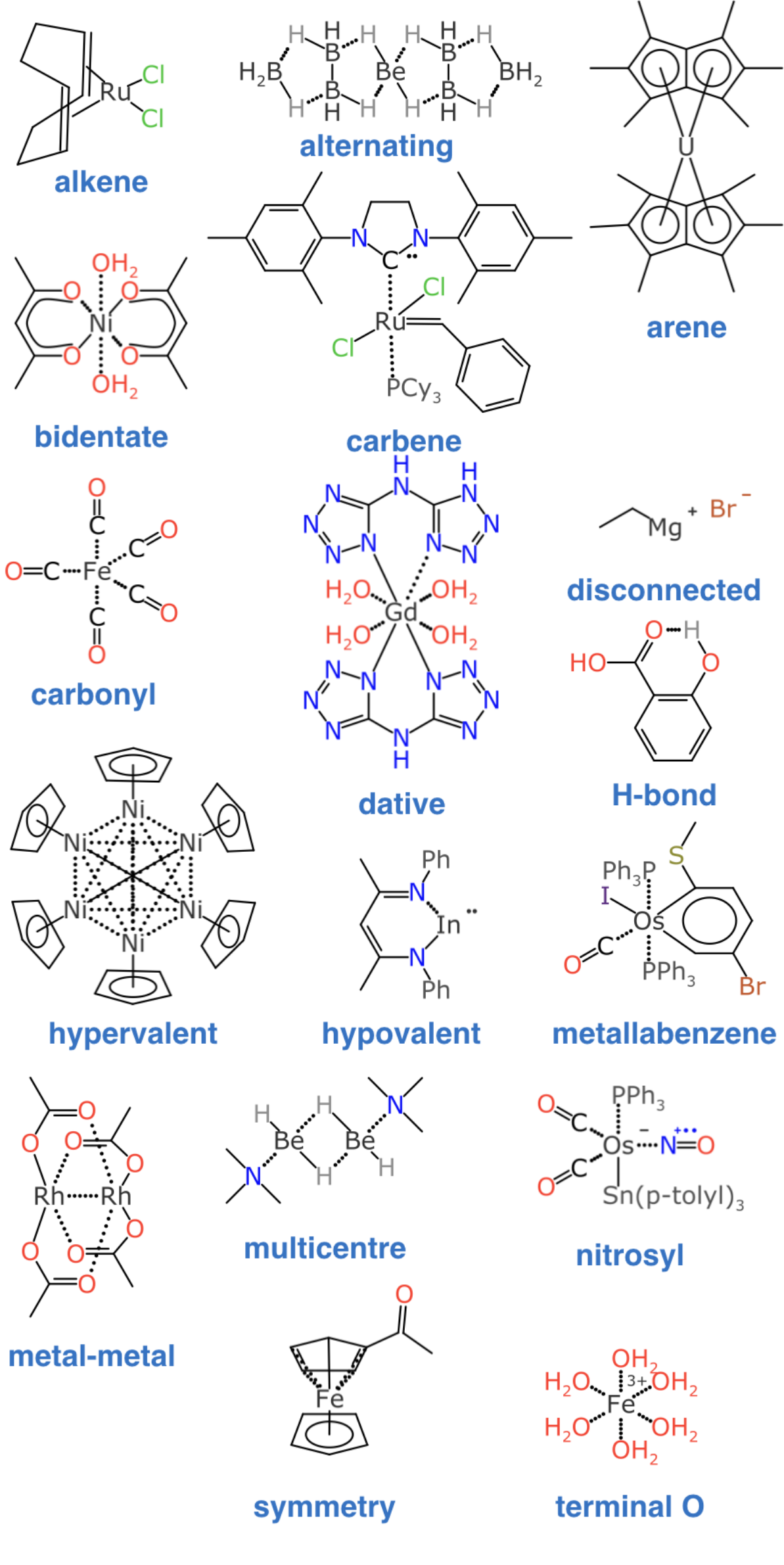
As the data gathering part of the project started to come together, I decided to try out some ideas about inorganic bonding informatics that I’ve been tinkering with for a long time, which involves dividing up the molecule into “dot paths”, or perhaps better described as electron delocalisation islands. The principle is that you start by identifying the subgraphs which are unified by are π-electrons or other kinds of facile mobility. Within each of those island subgraphs, it doesn’t matter where the input structure placed the bonds or charges, because they just readjust themselves: what matters is that you have the right number of electrons on each island. By averaging out these delocalised properties, it becomes possible to classify the atoms & bonds in a way that is consistent for a large variety of styles for representing the bonds, which is the core prerequisite of a canonical identifier.
The biggest caveat is that the atom & bond graph must be complete, and this includes the hydrogens that are normally left out of the input content, because people & the software that they use almost always assumes that they can be calculated, even though that is frequently not true. Until working on this project I had no useful external datasources to test out such ideas, but this all changed once I got access to the internal CSD representations, which take extremely great care to capture the graph completely and correctly.
The current state of this project is that now we have a thorough validation set of real world inorganic compounds, which demonstrates that the current standard InChI algorithm really does need to be improved; along with a proof-of-concept demonstration that it is possible to create a canonical ordering system that works for all of the examples, and which is remarkably robust to some quite bizarre drawing styles. Which means that chemists won’t have to draw their inorganic compounds any differently – they’ll just have to make sure the hydrogens are specified, and that they don’t get lazy about using undefined abbreviations.
This is great – sorely needed. I’m sure you know of some of the literature precedents (e.g. Dietz and particularly Gasteiger). I don’t know how much bandwidth I have at the moment, but let me know if I can help. I’ll certainly publicize the effort amongst computational inorganic chemists, who may have some comments or species to add to the mix.