 Since the Living Molecules app is now live on the iTunes AppStore, it’s time to say a little more about what its intended purpose is. While the potential uses of a “molecular QR code” system probably include a whole lot of things that I haven’t thought of, the original concept was inspired by the idea of adding supplementary information to posters.
Since the Living Molecules app is now live on the iTunes AppStore, it’s time to say a little more about what its intended purpose is. While the potential uses of a “molecular QR code” system probably include a whole lot of things that I haven’t thought of, the original concept was inspired by the idea of adding supplementary information to posters.
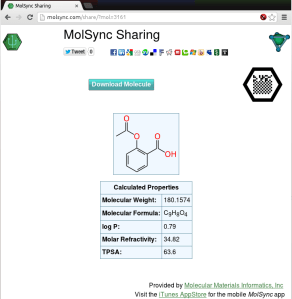
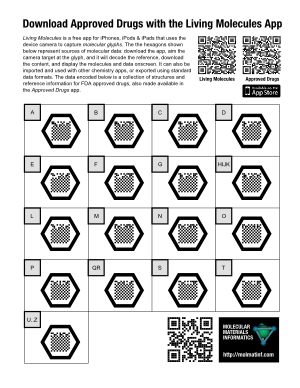
The example poster on the right (click for the PDF version) takes the concept of “supplementary” to an absurdity, but that’s OK, because it’s a demo poster made to prove a point: each of the 17 molecular glyphs corresponds to a portion of the molecule collection that makes up the raw data for the Approved Drugs app. The data is hosted on molsync.com, where it has been residing for some time (note that it can also be found in the Drug Repurposing topic for Open Drug Discovery Teams).
If you were to print out this Letter-sized poster and stick it up on a wall, anyone who walks up and takes a look at it will either: (1) recognise the molecular glyphs for what they are, whip out their iThing and open up their already installed copy of Living Molecules; or (2) read the explanatory text to find out what it’s all about, snap the regular QR code at the top to get to the iTunes AppStore to download the free app, then proceed as for (1); or (3) send me an angry email demanding that an Android version be made available immediately.
For a more realistic poster, which is describing actual science rather than a techno-utility, >95% of the surface area of the poster would be taken up with the usual things: title, attribution, feature pictures, captions, text, graphics, etc. A modestly sized molecular glyph might be found in a corner, with a little note mentioning something like “The actual data used to produce the structure-activity plots shown above is available by capturing this glyph“.