 Using chemical substructures with query annotations is a cheminformatics trick that goes back a long way: by annotating the query structure with additional logical constraints, the utility of this functionality can be juiced up, by matching a variety of certain kinds of fragments, rather than literal pieces. The amount of work involved to implement such a system is quite evenly divided between providing the low-level algorithm for doing the matching, and providing a high-level editor for people to design the queries. One of the “TO-DO” features in the XMDS beta version has taken some steps toward fulfilling the latter. Continue reading
Using chemical substructures with query annotations is a cheminformatics trick that goes back a long way: by annotating the query structure with additional logical constraints, the utility of this functionality can be juiced up, by matching a variety of certain kinds of fragments, rather than literal pieces. The amount of work involved to implement such a system is quite evenly divided between providing the low-level algorithm for doing the matching, and providing a high-level editor for people to design the queries. One of the “TO-DO” features in the XMDS beta version has taken some steps toward fulfilling the latter. Continue reading
Visualisation of structure-activity models: fudging it with a widget
 One of the opinions (arguably of the educated variety) that I’ve been pushing for awhile now is the idea that when a model building or visualisation technique requires a user parameter in order to get the correct result, that is essentially an admission of partial failure. If the method really was so great, then it would be able to figure it out, because a parameter is an extra degree of freedom that the method has punted on. Now of course this is not a rule by any stretch of the imagination, and there are numerous exceptions, or grey areas between what’s a parameter and what’s an integral component of the source data. But sometimes a parameter really is just something that a method ought to know, but gives up and passes the burden on to the user – and that’s not necessarily a bad thing, as long as we admit it. Continue reading
One of the opinions (arguably of the educated variety) that I’ve been pushing for awhile now is the idea that when a model building or visualisation technique requires a user parameter in order to get the correct result, that is essentially an admission of partial failure. If the method really was so great, then it would be able to figure it out, because a parameter is an extra degree of freedom that the method has punted on. Now of course this is not a rule by any stretch of the imagination, and there are numerous exceptions, or grey areas between what’s a parameter and what’s an integral component of the source data. But sometimes a parameter really is just something that a method ought to know, but gives up and passes the burden on to the user – and that’s not necessarily a bad thing, as long as we admit it. Continue reading
XMDS and Mac OS X integration: drag’n’drop and previews
 The OS X Molecular DataSheet (XMDS) app has been getting a bit more attention lately, after being interrupted by the summer conference season (slides, poster, slides & more slides) and the PolyPharma app (which is now submitted to the AppStore and waiting for approval). The next step that brings XMDS closer to being a finished product rather than a functional beta is deeper integration with OS X. Continue reading
The OS X Molecular DataSheet (XMDS) app has been getting a bit more attention lately, after being interrupted by the summer conference season (slides, poster, slides & more slides) and the PolyPharma app (which is now submitted to the AppStore and waiting for approval). The next step that brings XMDS closer to being a finished product rather than a functional beta is deeper integration with OS X. Continue reading
Some app housecleaning & ACS aftermath
 Writing this post from Boston as the American Chemical Society meeting winds down – by Wednesday evening most of the action is over – it has been a good one. My first talk (see slideshare) went without a hitch, which was not a foregone conclusion, since the second half involved a live demo of the PolyPharma app, which didn’t exist 2 weeks ago, is half finished, and consists mostly of brand new original code.
Writing this post from Boston as the American Chemical Society meeting winds down – by Wednesday evening most of the action is over – it has been a good one. My first talk (see slideshare) went without a hitch, which was not a foregone conclusion, since the second half involved a live demo of the PolyPharma app, which didn’t exist 2 weeks ago, is half finished, and consists mostly of brand new original code.
Besides a new app on the way and a busy month of conference presentations, there are changes afoot in the app lineup which will start to be noticed on the Molecular Materials Informatics product page shortly. For chemistry app users, there’s good news and bad news. The bad news is that some apps have been retired, and some more are slated for the glue factory in the near future. The good news is that all the rest of the flagship apps are now free, as of several days ago, and the intention is to leave them that way indefinitely. Continue reading
The PolyPharma app: a mash-up of ideas and technology
![]() Things have been a bit quiet around here lately, but there is a good reason: at the Gordon conference on Computer Aided Drug Design a couple of weeks ago, a confluence of ideas came together. With the opportunity to observe a series of talks and posters about what practitioners in the industry are interested in right now, and the chance to discuss my recent projects and gauge the level of interest, it all happened to coincide with a number of partially complete product explorations that have been sitting on the workshop floor, waiting for the right time and place to assemble release to the outside world. Continue reading
Things have been a bit quiet around here lately, but there is a good reason: at the Gordon conference on Computer Aided Drug Design a couple of weeks ago, a confluence of ideas came together. With the opportunity to observe a series of talks and posters about what practitioners in the industry are interested in right now, and the chance to discuss my recent projects and gauge the level of interest, it all happened to coincide with a number of partially complete product explorations that have been sitting on the workshop floor, waiting for the right time and place to assemble release to the outside world. Continue reading
What’s in a name: Molecular Materials
I am presently attending the Gordon Research Conference: Computer Aided Drug Design meeting, having a wonderful time, and at this moment taking an afternoon break to relax and catch up on some work. This is one of my favourite properties of the GRC meeting format, because it means that it’s not necessary to be pretty much out of the loop for the entire week.
So far I’ve met and reacquainted myself with more scientists of my own particular species than I can count, and a recurring theme of the introduction/renewal process is taking turns to explain what we’re both doing these days. One part of this which seems a little incongruous is that the name of the company that I created is called “Molecular Materials Informatics”. Since the occasion is a conference that is exclusively devoted to the pursuit of drug molecules and a few related parts of life sciences, this requires some explanation. Continue reading
Inline abbreviations with XMDS
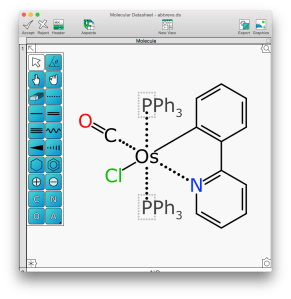 Abbreviations are a common shorthand used by chemists to alleviate the tedium of drawing out structures by hand, and in many areas – including much of organometallic and inorganic chemistry – use of abbreviations is more or less essential to achieve any kind of visual clarity. This presents a challenge for cheminformatics, though, because there is no universal dictionary for abbreviations: different research groups use different shorthand notations for their own nefarious purposes. Now the beta version of XMDS is catching up to MMDS by adding the ability to conveniently define inline abbreviations, which are readable to humans in diagram form, but also completely well defined behind the scenes for the benefit of machines. Continue reading
Abbreviations are a common shorthand used by chemists to alleviate the tedium of drawing out structures by hand, and in many areas – including much of organometallic and inorganic chemistry – use of abbreviations is more or less essential to achieve any kind of visual clarity. This presents a challenge for cheminformatics, though, because there is no universal dictionary for abbreviations: different research groups use different shorthand notations for their own nefarious purposes. Now the beta version of XMDS is catching up to MMDS by adding the ability to conveniently define inline abbreviations, which are readable to humans in diagram form, but also completely well defined behind the scenes for the benefit of machines. Continue reading
Closing the collaborative loop: Bayesian models with XMDS, CDD Vault and MMDS
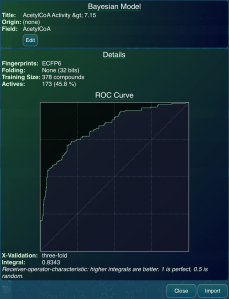 The CDD Vault web product has recently delivered on one of its promises: making models truly sharable to the community, including both open source and participating commercial products. Bayesian models can now be exported in a well documented and openly consumable format, which removes a major bottleneck in the creation and use of models for structure-activity predictions. Continue reading
The CDD Vault web product has recently delivered on one of its promises: making models truly sharable to the community, including both open source and participating commercial products. Bayesian models can now be exported in a well documented and openly consumable format, which removes a major bottleneck in the creation and use of models for structure-activity predictions. Continue reading
Always-on R/S and E/Z stereo labels with XMDS
 The most recent addition to the OS X Molecular DataSheet (XMDS) desktop app is calculation of stereochemistry labels as-you-edit, using the Cahn-Ingold-Prelog (CIP) designations (R/S, Z/E). The labelling can be switched off with a menu option, but since most people use software with its factory settings, this is more or less equivalent to having it hardcoded permanently. Continue reading
The most recent addition to the OS X Molecular DataSheet (XMDS) desktop app is calculation of stereochemistry labels as-you-edit, using the Cahn-Ingold-Prelog (CIP) designations (R/S, Z/E). The labelling can be switched off with a menu option, but since most people use software with its factory settings, this is more or less equivalent to having it hardcoded permanently. Continue reading
Aspects and XMDS: higher order metadata for datasheets
 Recent progress on the OS X Molecular DataSheet (XMDS) app, which is currently in beta, has involved the inclusion of aspects into the user interface for editing datasheets. The core format that the XMDS desktop app, as well as all the rest of the products from Molecular Materials Informatics, is the datasheet, which is a tabular format made up of rows and columns: like a spreadsheet, except that the columns are strongly typed, so you can’t just abuse it and put in whatever you want wherever you want. The minimalistic table datastructure can be supplemented by any number of aspects, which are directives that impose higher order layers of interpretation, which are often composed of multiple columns (e.g. a chemical reaction which cobbles together multiple molecules for its reactants, reagents and products). One of the fun software engineering challenges has been to make these complex representations fit seemlessly into the grid-based “spreadsheet” editor upon which XMDS operates. Continue reading
Recent progress on the OS X Molecular DataSheet (XMDS) app, which is currently in beta, has involved the inclusion of aspects into the user interface for editing datasheets. The core format that the XMDS desktop app, as well as all the rest of the products from Molecular Materials Informatics, is the datasheet, which is a tabular format made up of rows and columns: like a spreadsheet, except that the columns are strongly typed, so you can’t just abuse it and put in whatever you want wherever you want. The minimalistic table datastructure can be supplemented by any number of aspects, which are directives that impose higher order layers of interpretation, which are often composed of multiple columns (e.g. a chemical reaction which cobbles together multiple molecules for its reactants, reagents and products). One of the fun software engineering challenges has been to make these complex representations fit seemlessly into the grid-based “spreadsheet” editor upon which XMDS operates. Continue reading