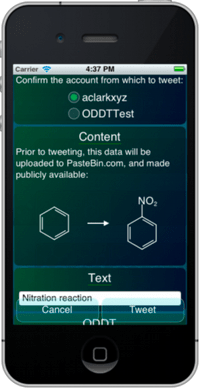
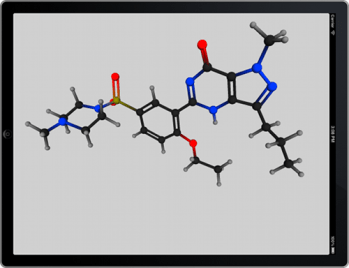
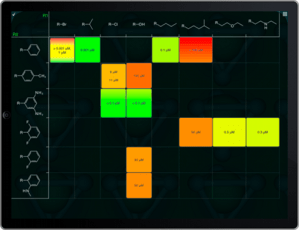
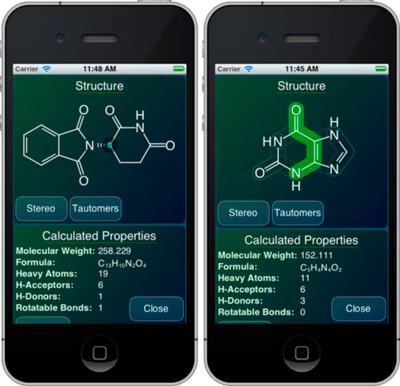
Now that my iPad 3 is delivered, unpacked and synced up, I can now officially declare that my last few reservations about Apple being a positive force in the technology industry have now vanished. I’ve been looking forward to the so-called retina-class display on a large-ish screen, and it’s everything I expected.
Everybody who has seen one of these agrees that yes, indeed, it’s pretty cool, but to someone who has spent more than two decades wrangling pixels, and experiencing each technology upgrade and grinding away on techniques for driving the improvements to their limit, this is one of the most exciting developments in a very long time. Put simply, these new displays mean that pixels don’t matter anymore. All those hours we spent integer-aligning widgets, optimising anti-aliasing algorithms, dithering, keeping multiple copies of bitmaps for screen/web/print quality, making icon and sprite design compromises to work around clarity…
… all of that now gone. Now our apps can look as good as a glossy magazine, and it’s even less effort than it was before.